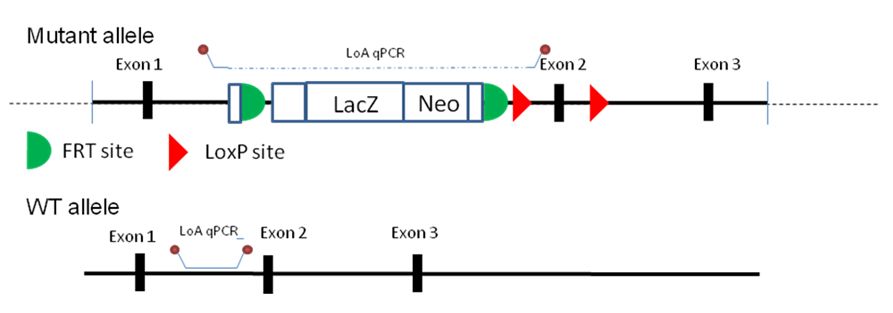
The wild type loss of allele (WTLoA) qPCR assay uses a hydrolysis probe assay (eg Applied Biosystems TaqMan technology) to determine the copy number of the wild type allele in a sample. Homozygotes will show no amplification, heterozygotes one copy and wild type mice will two copies. This is the inverse to what is seen on the Neo count assay. Primers and the hydrolysis probe are designed preferably in the region of the genome deleted during the mutant allele design phase (between the U5 and U3 sequences) although some flanking sequence can be used if necessary. The mutant allele will then either produce a product too large to be amplified by the PCR conditions or not contain the sequence at all, resulting in no amplification of product. Assays are usually designed to amplify a product of less than 100bp as this helps to attain maximum efficiency of the PCR reaction which is important for the downstream analysis and subsequent copy number determination (WTSI uses the 2-ddCt method for analysis) Care must be taken to avoid designing the assay in heavy repeat regions when possible as this may affect the assay efficiency and the subsequent copy number detection The theory behind the calling method is discussed further here and here: The LoA assay can detect a correct cassette insertion event in heterozygotes as well as in homozygotes which makes it a good QC tool for the F1 chimera progeny to confirm the targeting.